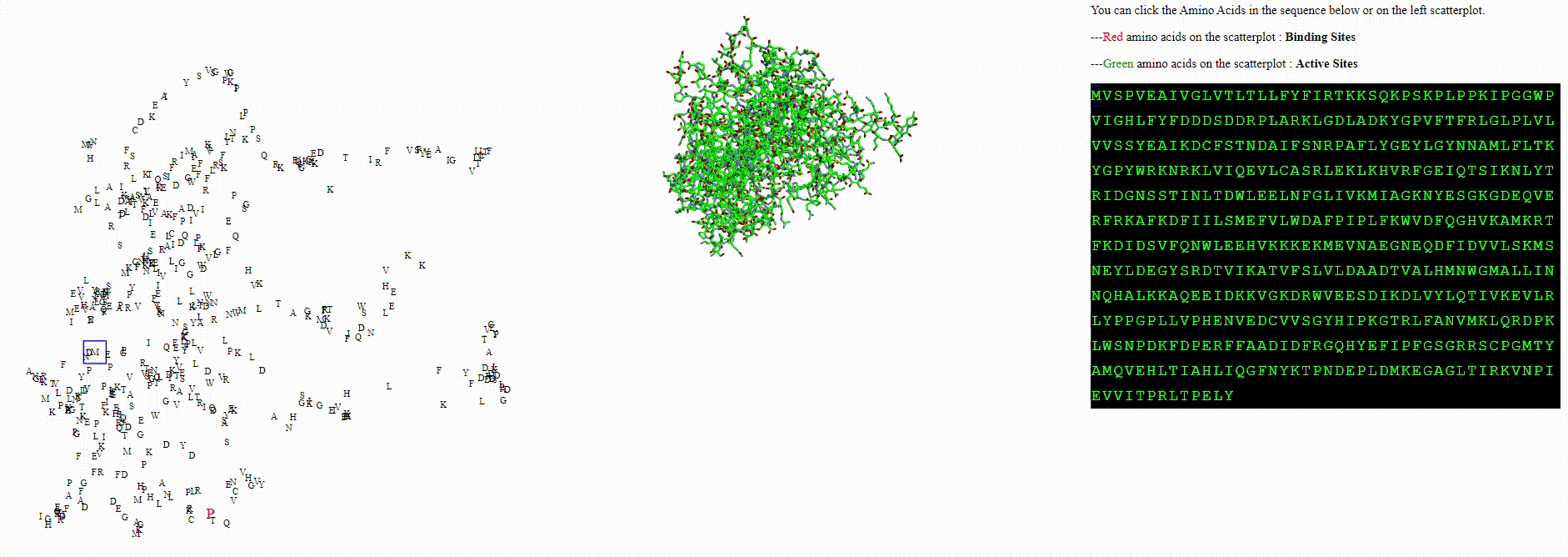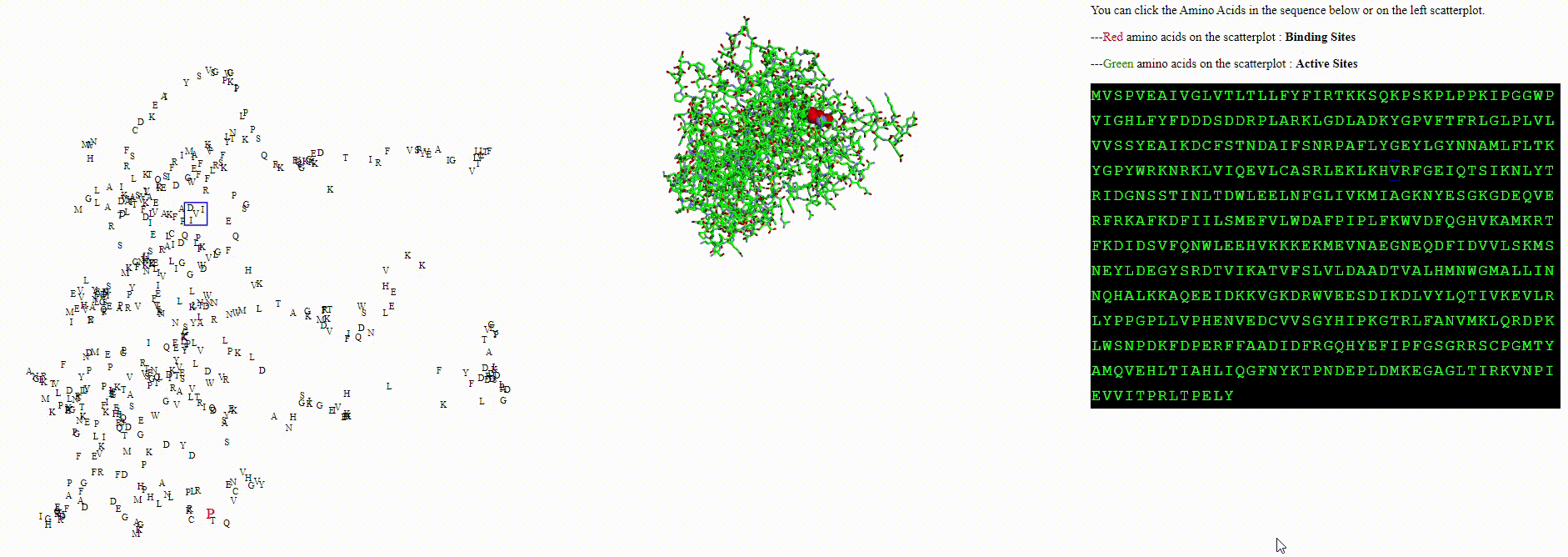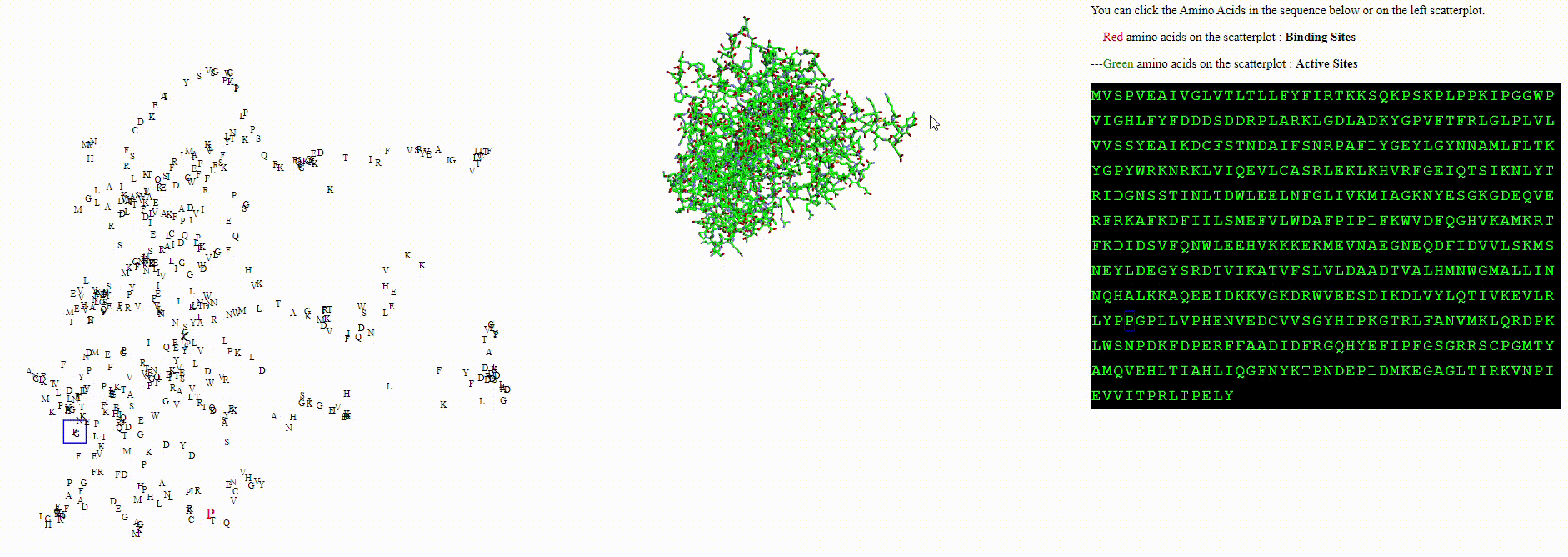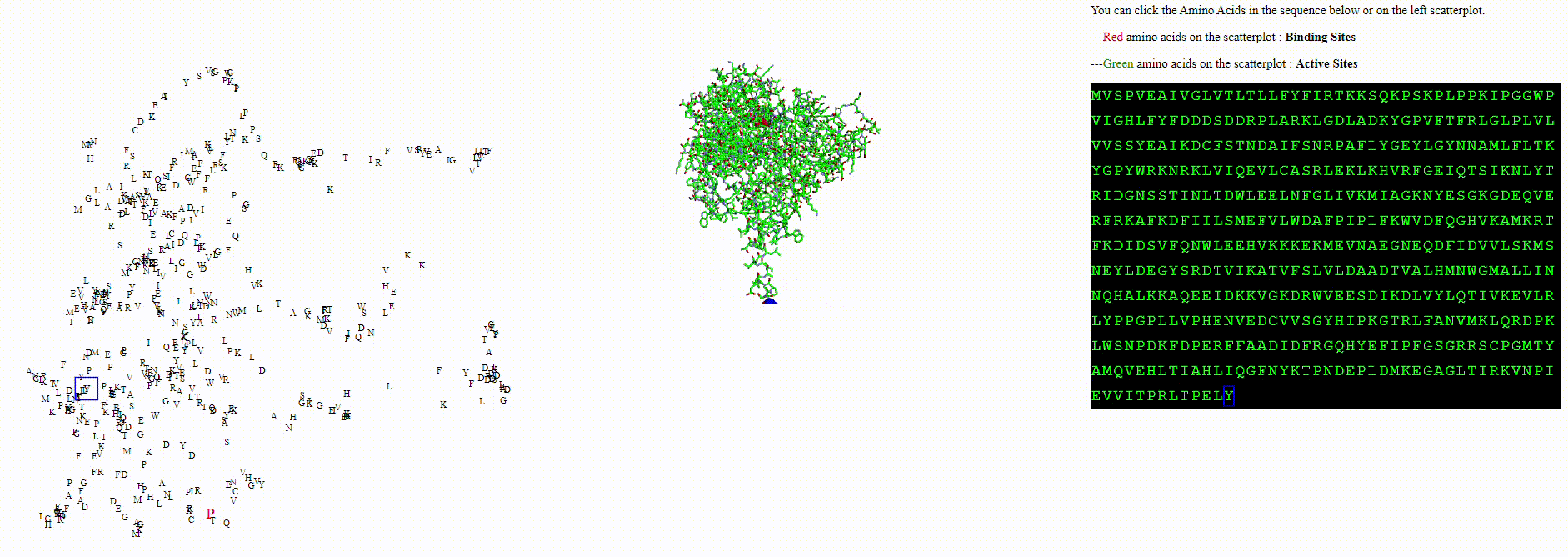
A protein biodiversity viewer with Deep Learning embeddings
This application was designed with a singular goal: to visualize amino acid embeddings within a given protein. Previously, I employed image-based machine learning models to embed and visualize unique patterns within each image, as well as clusters of images based on shared features. This same concept can be extended to the realm of proteins.
Language models like Meta’s ESM2 (Evolutionary Scale Modeling) transform each protein’s amino acid sequence into a corresponding token sequence. Amino acids are assigned unique IDs (e.g., Tyrosine = 15).
The protein sequence ‘ACZGLLL’ is converted to [0(A) 3(C) 12(Z) 4(G) 9(L) 9(L) 9(L)].
The machine learning model learns a pattern for each token. Intriguingly, the model can differentiate between the multiple Leucine (L) amino acids. Each token’s embedding incorporates its identity (Leucine) and the context of its neighboring amino acids. Any change in neighbors or their positions will modify all token embeddings. This core principle of context-awareness is what drives the power of language models (found in many of today’s famous AI systems).
Theoretically, this raises fascinating questions about how these amino acid chains are represented within a model trained on vast amounts of protein data. Some key questions include:
- How different are the embeddings for the same amino acid within a single protein (e.g. all the Lysine amino acids)?
- Are amino acids with features (like phosphorylation, or an active site) represented differently within the language model?
- How important is the size of the model for each amino acid’s representation?
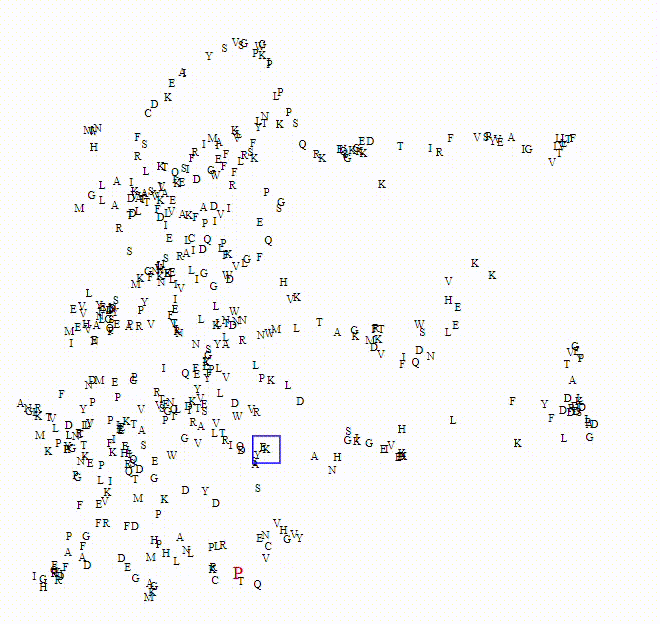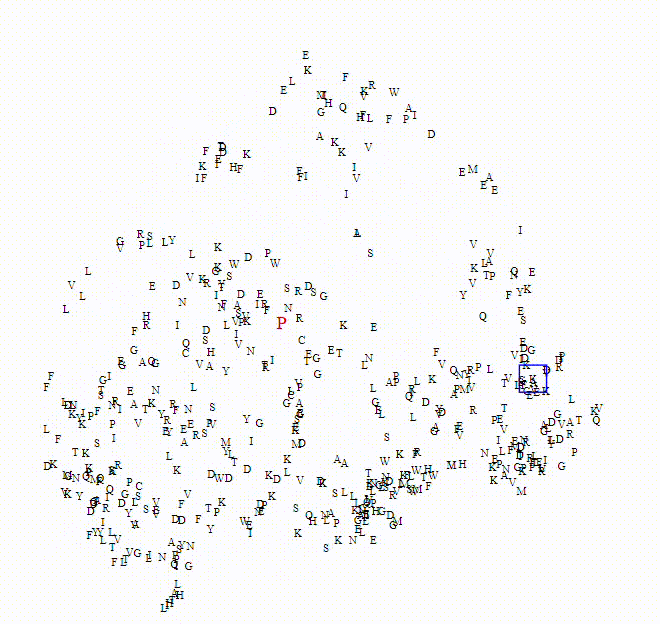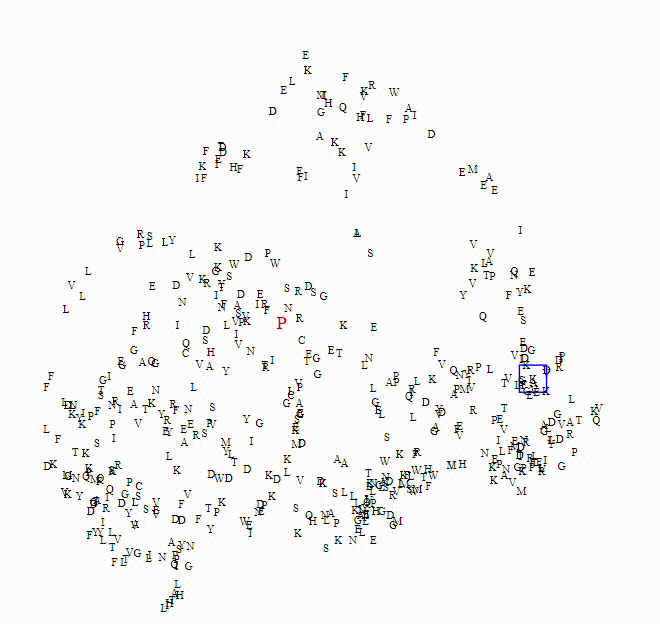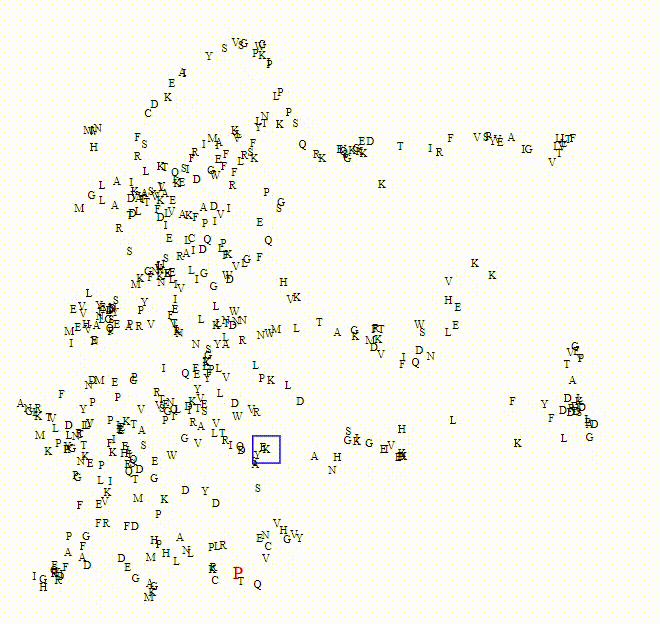
To explore these questions, I aimed to create an interactive visualization for analyzing this data. For a single protein, I would use an ESM model to calculate amino acid embeddings, apply dimensionality reduction, and then plot each amino acid on a scatterplot.
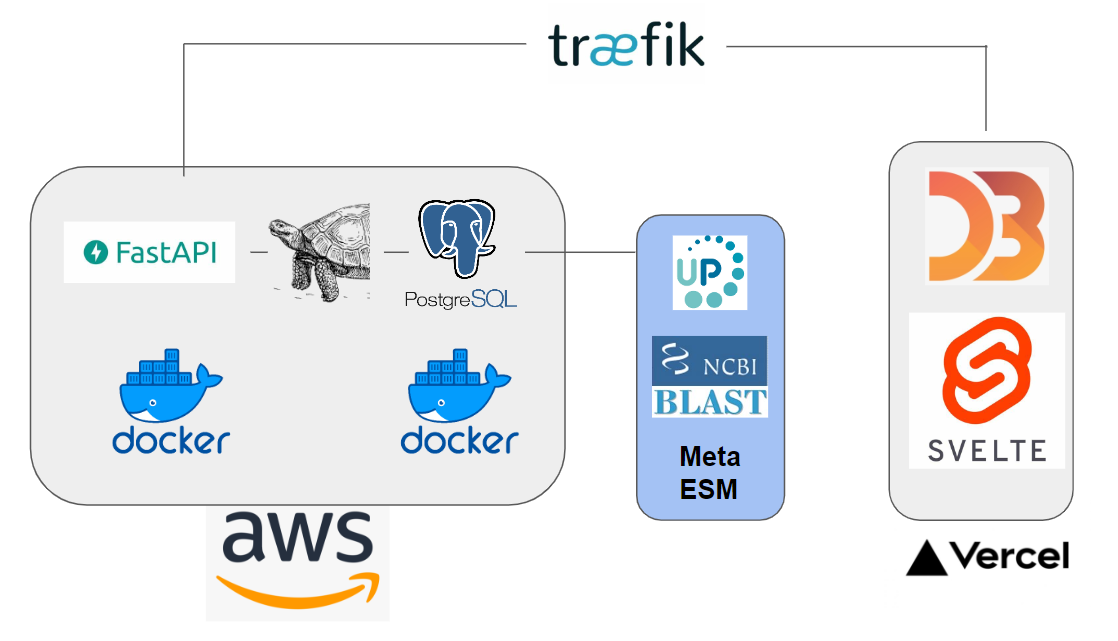
This project encompasses scraping UniprotKB, designing and populating a database (PostgreSQL), utilizing SQLAlchemy as an ORM, and employing Svelte and D3 for data visualization. Cloud hosting is provided by AWS, with Docker managing the backend, frontend, and database. PyTorch facilitates the use of ESM models.
Here we can see the embeddings transition between a small model (650M parameters) to a larger model (6B parameters)