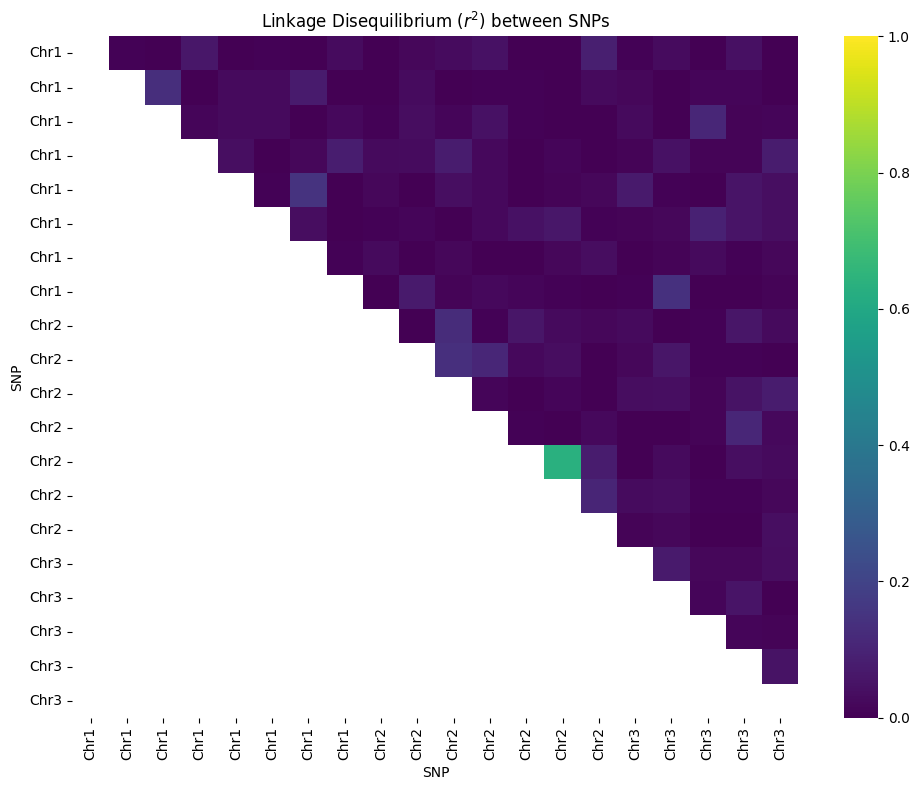
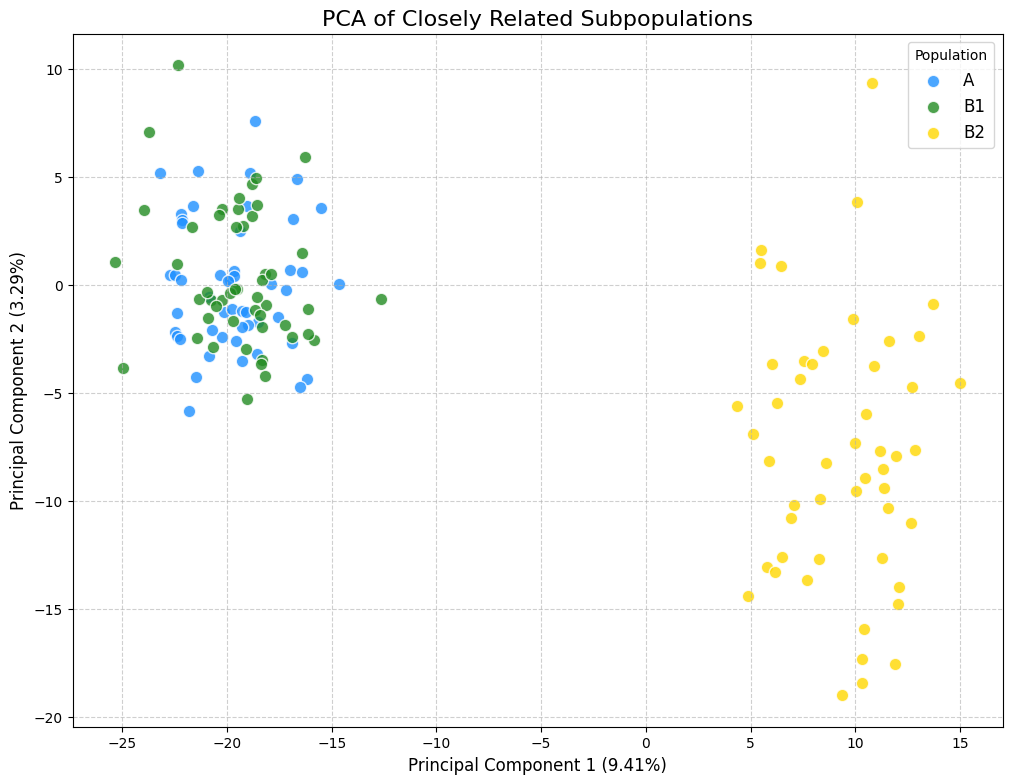
--- Genetic Map and Haplotypes for 3 Chromosomes ---
Genetic Map (Chr, Position):
[(np.int64(1), 148574), (np.int64(1), 161156), (np.int64(1), 168134), (np.int64(1), 230861), (np.int64(1), 296372), (np.int64(1), 404475), (np.int64(1), 458306), (np.int64(1), 526768), (np.int64(1), 720381), (np.int64(1), 1038824), (np.int64(1), 1479145), (np.int64(2), 61949), (np.int64(2), 426043), (np.int64(2), 697355), (np.int64(2), 779976), (np.int64(2), 877479), (np.int64(2), 900323), (np.int64(2), 913296), (np.int64(3), 337812), (np.int64(3), 419516)]
Individual 0:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 1:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 2:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 3:
Haplotype 1: [np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 4:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0)]
Individual 5:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 6:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0)]
Individual 7:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 8:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0)]
Haplotype 2: [np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0)]
Individual 9:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0)]
Individual 10:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 11:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 12:
Haplotype 1: [np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 13:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0)]
Individual 14:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1)]
Individual 15:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0)]
Individual 16:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Individual 17:
Haplotype 1: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(1)]
Individual 18:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(0)]
Individual 19:
Haplotype 1: [np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]
Haplotype 2: [np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(1), np.int32(0), np.int32(1), np.int32(1), np.int32(0), np.int32(0), np.int32(0), np.int32(0), np.int32(0)]